 |
 |
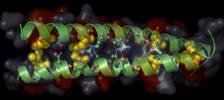 |
|
|||||||||||||
|
|
| Pfam ID | Description | Accession | Interpro ID | GO ID | ||||
| HTH_AraC | Bacterial regulatory helix-turn-helix proteins, AraC family | PF00165 | IPR000005 |
|
HTH_AraC
forms interactions with
| Pfam ID | Description | Accession | Interpro ID | GO ID | Notes | Source | |||||
| HTH_AraC | Bacterial regulatory helix-turn-helix proteins, AraC family | PF00165 | IPR000005 |
|
 |




|
|||||
| Ada_Zn_binding | Metal binding domain of Ada | PF02805 | IPR004026 |
|
 |

|
|||||
| RNA_pol_A_CTD | Bacterial RNA polymerase, alpha chain C terminal domain | PF03118 | IPR011260 |
|
 |


|
|||||
| AraC_E_bind | Bacterial transcription activator, effector binding domain | PF06445 | IPR010499 |
|
 |




|
|||||
| Response_reg | Response regulator receiver domain | PF00072 | IPR001789 |
|
 |

|
|||||
| DNA_binding_1 | 6-O-methylguanine DNA methyltransferase, DNA binding domain | PF01035 | IPR014048 |
|
 |


|
|||||
| DJ-1_PfpI | DJ-1/PfpI family | PF01965 | IPR002818 |
|
 |


|
|||||
| AraC_binding | AraC-like ligand binding domain | PF02311 |
|
 |


|
||||||
| HATPase_c | Histidine kinase-, DNA gyrase B-, and HSP90-like ATPase | PF02518 | IPR003594 |
|
 |

|
|||||
| Peripla_BP_1 | Periplasmic binding proteins and sugar binding domain of the LacI family | PF00532 | IPR001761 |
|
 |

|
|||||
| HhH-GPD | HhH-GPD superfamily base excision DNA repair protein | PF00730 | IPR003265 |
|
 |

|
|||||
| Peripla_BP_2 | Periplasmic binding protein | PF01497 | IPR002491 |
|
 |

|
|||||
| Methyltransf_1N | 6-O-methylguanine DNA methyltransferase, ribonuclease-like domain | PF02870 | IPR008332 |
|
 |

|
|||||
| AlkA_N | AlkA N-terminal domain | PF06029 | IPR010316 |
|
 |

|