 |
 |
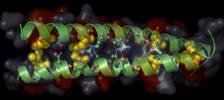 |
|
|||||||||||||
|
|
| Pfam ID | Description | Accession | Interpro ID | GO ID | ||
| PTS-HPr | PTS HPr component phosphorylation site | PF00381 | IPR005698 |
|
PTS-HPr
forms interactions with
| Pfam ID | Description | Accession | Interpro ID | GO ID | Notes | Source | |||||
| PTS_EIIA_1 | phosphoenolpyruvate-dependent sugar phosphotransferase system, EIIA 1 | PF00358 | IPR001127 |
|
 |




|
|||||
| PTS_EIIA_2 | Phosphoenolpyruvate-dependent sugar phosphotransferase system, EIIA 2 | PF00359 | IPR002178 |
|
 |




|
|||||
| PTS-HPr | PTS HPr component phosphorylation site | PF00381 | IPR005698 |
|
 |



|
|||||
| PEP-utilizers | PEP-utilising enzyme, mobile domain | PF00391 | IPR008279 |
|
 |



|
|||||
| Peripla_BP_1 | Periplasmic binding proteins and sugar binding domain of the LacI family | PF00532 | IPR001761 |
|
 |


|
|||||
| EIIA-man | PTS system fructose IIA component | PF03610 | IPR004701 |
|
 |



|
|||||
| PEP-utilisers_N | PEP-utilising enzyme, N-terminal | PF05524 | IPR008731 |
|
 |




|
|||||
| Hpr_kinase_C | HPr Serine kinase C-terminus | PF07475 | IPR011104 |
|
 |


|
|||||
| V-set | Immunoglobulin V-set domain | PF07686 | IPR013106 |
|
 |

|
|||||
| PEP-utilizers_C | PEP-utilising enzyme, TIM barrel domain | PF02896 | IPR000121 |
|
 |


|